Stories in Science Special Series
Growing Up in Science: David M. Schneider
David Schneider: “When I finished my masters, I applied to 11 PhD programs and was rejected by all of them. The next obvious step (to me) was to cold-call the director of graduate studies at Columbia (where I had just been rejected) and ask if there were any labs that would hire me for a year.”

David M. Schneider
[su_boxbox title=”About”]Dr. Schneider earned a bachelor’s degree in electrical engineering with a focus on control systems and prosthetics. During his undergraduate years, he spent two summers as an intern at a medical device company, where he holds a patent for the software and hardware he developed for building intelligent pacemakers. David then earned a master’s degree in biomedical engineering, where he used electrophysiology, behavior, and computational modeling to study how barn owls hunt their prey. After finishing his M.S., David spent 11 months as a research assistant at Columbia University. During that short time he performed experiments leading to 3 manuscripts on primate behavior, vision and attention. The following year David continued at Columbia as a graduate student, rotating through three labs, one of which resulted in a first author paper. David joined the lab of Sarah Woolley as its first graduate student, where he published 8 more papers (3 first author) and received multiple fellowships and awards, including an NRSA. David moved to Duke University for a postdoc in 2012 and published his first 2 papers within 24 months. As a postdoc he was the recipient of a Helen Hay Whitney Foundation postdoctoral fellowship and an Allison Doupe Fellowship from the McKnight Foundation. David does not have a K99/R00, but only because he declined it (after receiving a perfect score of 10) to accept a Career Award from the Burroughs Wellcome Fund. David started his lab at NYU in January of 2018 and was recently named a Searle Scholar. The story below is re-published in collaboration with Growing up in science. Learn more about the Schneider Laboratory. The story is co-published in collaboration with Growing up in Science. [/su_boxbox]
[dropcap]I[/dropcap] grew up in rural North Dakota. I was a mediocre student in high school and applied to exactly one college, North Dakota State University, where it took me 5 years to earn a bachelor’s degree in electrical engineering. It took 5 years because I nearly flunked out in years 2 and 3 and rarely attended class except in years 1 and 5. Despite bad grades and a broken compass, I applied to 4 masters programs and got into 1, for the sole reason that the chair at that department was a former professor at NDSU.

David M. Schneider
When I finished my masters, I applied to 11 PhD programs and was rejected by all of them. The next obvious step (to me) was to cold-call the director of graduate studies at Columbia (where I had just been rejected) and ask if there were any labs that would hire me for a year. There luckily was, and so I packed my bags and moved to New York. I worked at Columbia for a year, reapplied to grad school and was accepted everywhere I applied this time. After starting my PhD, I naively interacted with PIs as if they were peers. This includes the time I bumped into Richard Axel at a bodega the summer before grad school and told him I was going to join his lab for his first rotation. I did indeed rotate in the Axel lab.
During my first year in grad school, I transiently suffered from anxiety attacks, which just about made me drop out of grad school. But I recovered and ultimately ended up studying bird brains with Sarah Woolley, then moved to Duke for a postdoc before coming back to NYC in 2018. My path eventually worked out because I married someone much smarter than myself and because rather than work 80-hour weeks, I spend most of my evenings and weekends hanging out with my children.
Cover image by Gerd Altmann from Pixabay
Metrics
Sessions
Total number of Sessions. A session is the period time a user is actively engaged with the page.
Visitors
Users that have had at least one session within the selected date range. Includes both new and returning users.
Page views
Pageviews is the total number of time the article was viewed. Repeated views are counted.

-
 Audio Studio1 month ago
Audio Studio1 month ago“Reading it opened up a whole new world.” Kim Steele on building her company ‘Documentaries Don’t Work’
-
Civic Science Observer1 week ago
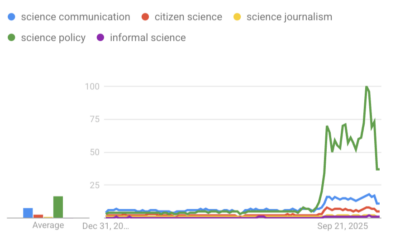
‘Science policy’ Google searches spiked in 2025. What does that mean?
-
Civic Science Observer1 month ago
Our developing civic science photojournalism experiment: Photos from 2025
-
Civic Science Observer1 month ago
Together again: Day 1 of the 2025 ASTC conference in black and white
Contact
Menu
Designed with WordPress