Stories in Science Special Series
Getting Started in Academia
Cristina Savin: “In retrospect, I had very little idea what I was doing. After the first few rejections, I started doubting that I belonged in academia at all.”

Cristina Savin
[su_boxbox title=”About”]Prof. Cristina Savin received her PhD from Goethe University in Frankfurt, Germany, after doing theoretical work on the computational roles of different forms of plasticity in the group of Jochen Triesch at the Frankfurt Institute for Advanced Studies. She then moved to Cambridge University for a postdoc with Mate Lengyel, followed by a short research visit at ENS in Paris in the group of Sophie Deneve, and an independent research fellowship at IST Austria, working in collaboration with Gasper Tkacik and Joszef Csicsvari. Since 2017 she has been an Assistant Professor in the Center for Neural Science and the Center for Data Science at NYU. Her research sits at the intersection between neuroscience and machine learning, with a focus on learning at the level of circuits. Her group develops both theory and new data analysis tools for understanding how neural circuits do useful computation, in collaboration with several experimental partners. Cristina has also recently started an industry collaboration centered on nervous system computer interfaces for medical applications. Learn more about the Savin Lab. The story is co-published in collaboration with Growing up in Science. [/su_boxbox]
[su_boxnote note_color=”#d5ecb3″]Some Take Aways
- Give yourself time to figure out what interests you.
- Don’t feel too proud/embarrassed to ask for help.
- Be proud of your achievements but learn to also appreciate the mistakes made along the way.[/su_boxnote]

Dr. Cristina Savin
[dropcap]G[/dropcap]rowing up in a small town in Transylvania, my vision of possible career options was quite narrow. As someone who excelled academically but felt no passion for any particular subject (beyond reading indiscriminately and painting), I toyed with the idea of going to medical school before finally ending up, almost by chance, as a computer science (CS) major in high-school and then at the Technical University in Cluj-Napoca. There, I fell in love with both CS and university life; I continued to be top of the class while still painting and generally doing more things that should realistically fit in 24 hours.
In my third year, I briefly worked in industry as a software engineer, which helped confirm my choice of staying in academia. In Romania, this would usually mean staying on at the same university and working slowly through the ranks towards a permanent position. The Dean, one of my early mentors, pushed me to go abroad for my PhD instead. After some complicated family negotiations, I accepted a PhD position in Frankfurt, working with Jochen Triesch on computational roles of neuroplasticity. Despite struggling with living abroad for the first time and a rather unwelcoming environment, research proceeded relatively smoothly, resulting in several publications and one contributed Cosyne talk.
I handed in my thesis after three and a half years. While the questions I was asking during my PhD remained prominent in my mind, by the end of my stay in Frankfurt, I was feeling dissatisfied with the methodology. Prompted by a collaboration with Joerg Luecke, at the time a postdoc at the same institute, I started shifting towards machine learning based approaches to studying brain computation. Why? Well, traditional approaches involved too much trial and error and figuring with model parameters. The machine learning methods offered a neat mathematically clean way of thinking. They have their own limits which I eventually found out but machine learning provided a more natural way for me to think about computation in general and brain computation in particular.
I sat for the first time in an experimental lab and quickly discovered that none of the neuroscientists around were interested in talking to me.
I landed my dream postdoc in Cambridge University working in the lab of Mate Lengyel in collaboration with Peter Dayan. It quickly became apparent that I had a great deal to learn, but I persevered and managed to develop several interesting ideas, with corresponding publications.
While external validation that I was on the right track was hard to come by, I did manage to get a selective Neural Information Processing Systems (NIPS) talk, and after two and a half years, I felt I was in a good position to go back on the job market. I had multiple offers, but eventually decided on an independent research fellowship in Vienna as I wanted to focus more on data analysis. I ended up deferring the start date to spend some time in Paris in the lab of Sophie Deneve to work on what became one of her first independent lines of research (which brought another NIPS spotlight and a second Cosyne talk).
In Vienna, I sat for the first time in an experimental lab and quickly discovered that none of the neuroscientists around were interested in talking to me. It took a few months of participating in journal clubs and research talks until finally someone came to me with a question, and I finally got a stamp of approval when one of my suggestions turned into an actual experiment. In parallel, I started looking into faculty jobs, initially restricting the search to Europe but eventually also sending a few applications in the US, despite strong family opposition. I struggled with understanding how the job marked worked, especially in the US; lack of mentorship partly because of me being too stubborn and proud to ask for help; and more generally figuring out the best way to pitch my research to a broad audience. It took a while before starting to get invited for interviews; even longer for an actual offer to materialize. In retrospect, I had very little idea what I was doing. After the first few rejections, I started doubting that I belonged in academia at all.
Fortunately, chance intervened. At the point when I was ready to give up, I received an email encouraging her to apply for a joint CNS-CDS position at NYU. Long story short, I eventually was offered the job. Getting started was not easy, but things are starting to come together thanks to some rather amazing students. I am excited about starting several new collaborations and developing new grad courses. I am excited about the challenges ahead.
Metrics
Sessions
Total number of Sessions. A session is the period time a user is actively engaged with the page.
Visitors
Users that have had at least one session within the selected date range. Includes both new and returning users.
Page views
Pageviews is the total number of time the article was viewed. Repeated views are counted.
The CS Media Lab is a Boston-anchored civic science news collective with local, national and global coverage on TV, digital print, and radio through CivicSciTV, CivicSciTimes, and CivicSciRadio. Programs include Questions of the Day, Changemakers, QuickTake, Consider This Next, Stories in Science, Sai Resident Collective and more.

-
 Audio Studio1 month ago
Audio Studio1 month ago“Reading it opened up a whole new world.” Kim Steele on building her company ‘Documentaries Don’t Work’
-
Civic Science Observer1 week ago
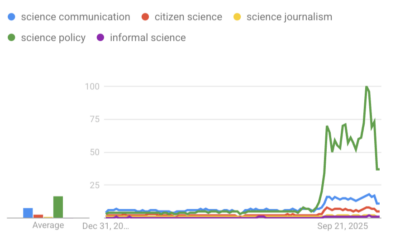
‘Science policy’ Google searches spiked in 2025. What does that mean?
-
Civic Science Observer1 month ago
Our developing civic science photojournalism experiment: Photos from 2025
-
Civic Science Observer1 month ago
Together again: Day 1 of the 2025 ASTC conference in black and white
Contact
Menu
Designed with WordPress